Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
Signaling Pathways Project Analysis Generates Hypotheses Around Host Responses to SARS-CoV-2 Infection in COVID-19
How were Signaling Pathways Project (SPP) analytics applied to characterize signaling pathways involved in SARS-CoV-2 - host interactions?
Development of coronavirus infection transcriptional regulatory networks from publicly archived datasets
To help catalyze the development of novel anti-coronavirus (CoV) therapeutics by the research community, Dr. McKenna’s SPP group generated consensomes to identify human genes that were most consistently induced or repressed in response to infection of cells by the three major CoVs: Middle East respiratory syndrome coronavirus (MERS) and severe acute respiratory syndrome coronaviruses SARS-CoV-1 (SARS1) and SARS-CoV-2 (SARS2, which causes COVID-19). SPP then computed the consensomes against a library of transcriptomic and ChIP-Seq high confidence transcriptional targets to identify signaling nodes whose gain or loss of function accompanies infection by CoVs.
Validation of consensomes
Validating the predictive power of the consensome with respect to clinical COVID-19, a 12-gene signature shown to correlate with COVID-19 severity in a recent Nature paper was robustly overrepresented among genes that were consistently induced in response to SARS2 infection (Fig. 1). Validating the signaling node analysis, Figure 2 shows ranked gene essentiality Z scores from a recent genome-wide CRISPR screen for host factors with roles in SARS2. Red genes represent those with essentiality Z-scores greater than 2. Shown in yellow are nodes predicted by SPP analytics to have a role in the transcriptional response to SARS2 infection (Q<1E-5). Confirming the biological relevance of SPP predictions, these Q<1E-5 nodes are robustly over-represented amongst genes with CRISPR essentiality Z scores >2.
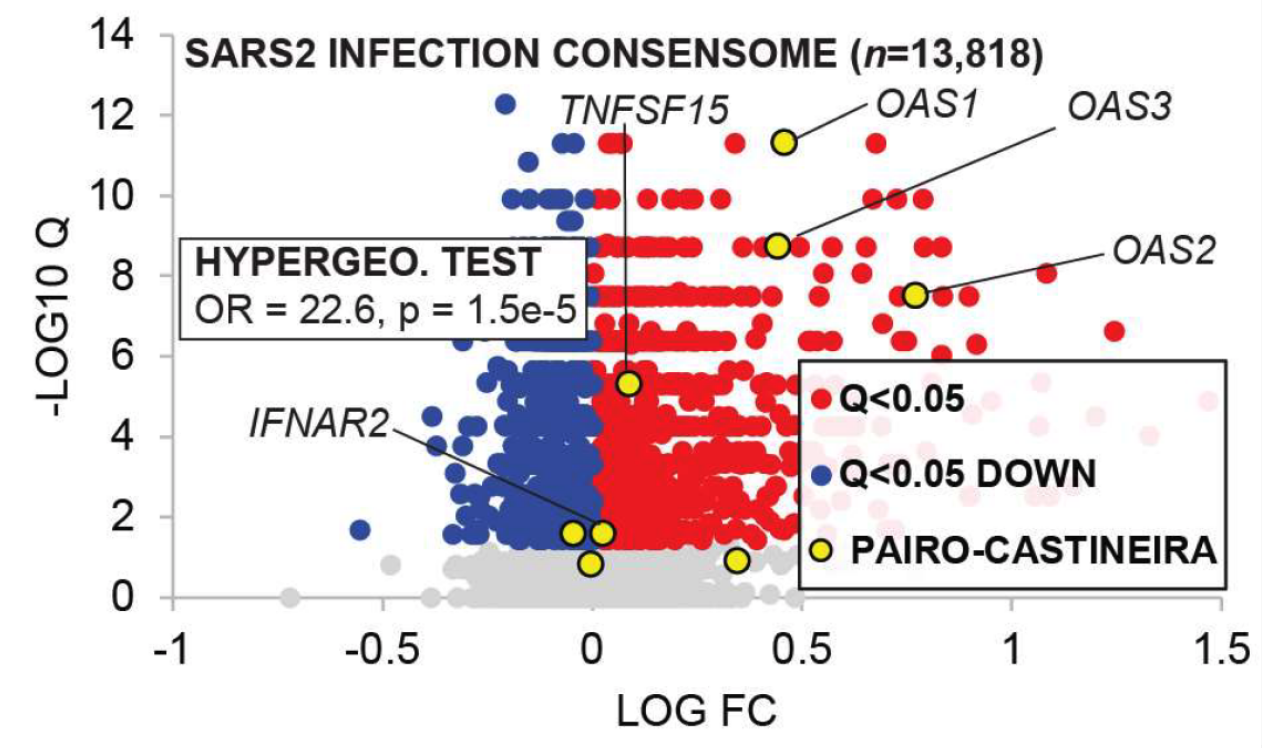
Figure 1
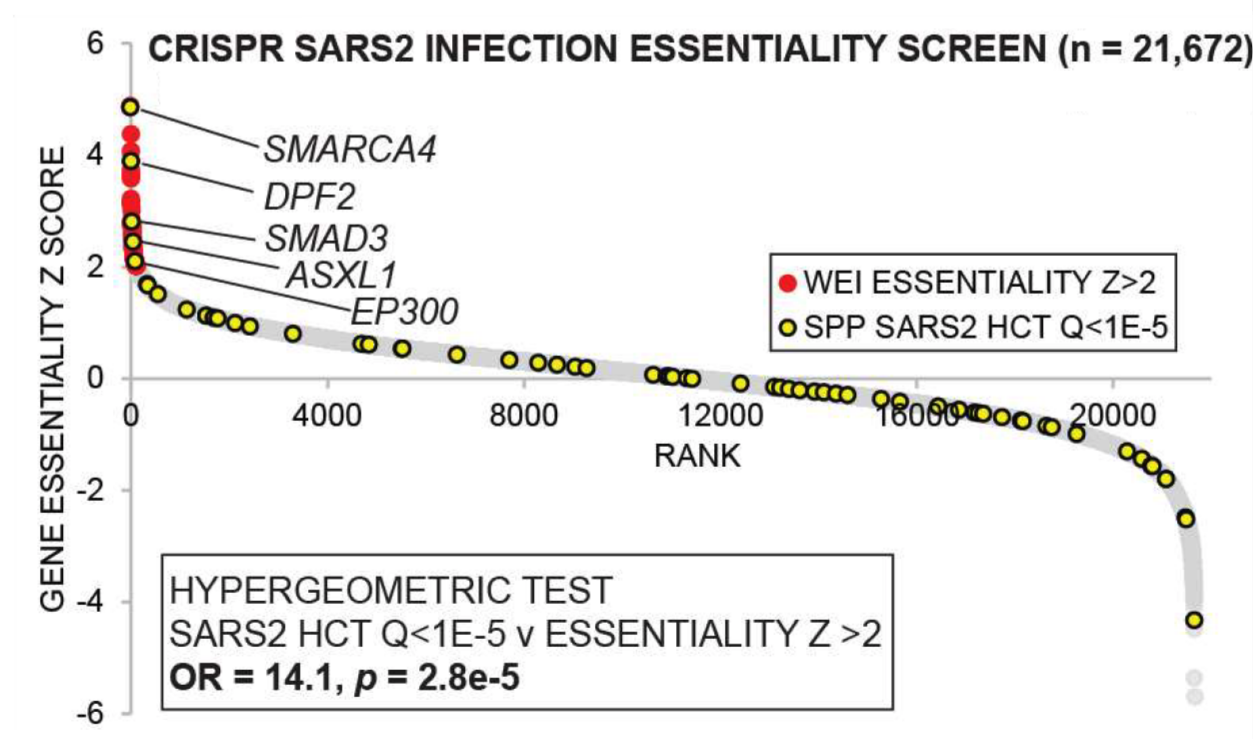
Figure 2
Data mining and visualization using SPP and NDEx
To enhance access to the SARS-CoV-2 networks by COVID-19 researchers, Dr. McKenna’s group collaborated with the Network Data Exchange repository (NDEx) at the University of California San Diego, to make the consensome analyses available as intuitive Cytoscape-style networks. Because they do not require bioinformatics training to manipulate, the SARS-CoV-2 Cytoscape networks allow bench researchers to more easily mine the datasets to generate or validate research hypotheses.
Generation of novel hypotheses around CoV-host interactions
SPP analytics generated numerous in silico hypotheses for cellular signaling pathways impacted by SARS-CoV-2 infection that were confirmed by subsequent independent wet bench experiments. Figure 3A shows a confidence heat map for selected nodes with SPP-predicted roles in SARS-CoV-2 infection. Numerous use cases validate these predictions: A CRISPR screen showed that numerous nodes with SPP-predicted roles in the response to SARS-CoV-2 infection were gene hits for SARS-CoV-2 resistance (SMARCA4, DPF2, ARID1A, LDB1, CREBBP,) and sensitization (BPTF and NONO; Fig. 3B). Consistent with predictions of roles for DNMT3A and TET2 in SARS-CoV-2 infection, there is a high frequency of DNMT3A and TET2 mutations in hospitalized COVID-19 patients (Fig. 3C). Reflecting significant intersection with BCL6 HCTs, the proportion of Bcl6+ B cells is greatly reduced in lymph nodes from COVID-19 patients relative to non-COVID-19 patients (Fig. 3D). Reflecting a predicted role for NELFE, phosphorylation of NELFE increases with duration of SARS-CoV-2 infection (Fig. 3E). Consistent with prediction of STAT2, a protective role of Stat2 in the reponse to infection is indicated by elevated SARS-CoV-2 viral titers in the lungs of Stat2 null Syrian golden hamsters relative to WT animals (Fig. 3F). Reflecting a predicted role for BRD4, SARS-CoV-2 transmembrane E protein has been shown to bind to BRD2 and BRD4 (Fig. 3G). Finally, SPP predicted that the SARS-CoV-2 infection impacts the function of CDK kinases, which are well characterized regulators of DNA replication and the cell cycle. Consistent with this, SARS-CoV-2 infection has been shown to be negatively correlated with profiles of cells in mitosis (Fig. 3H).
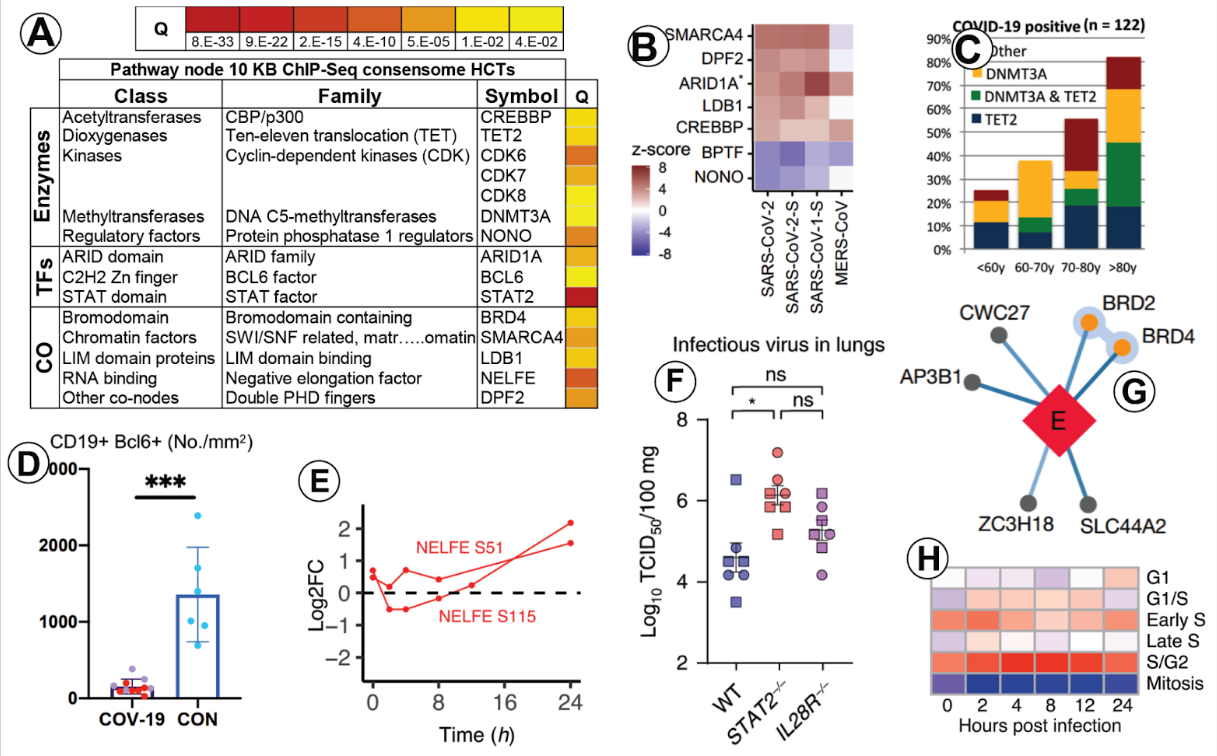
Figure 3
What is the tool(s) used:
Signaling Pathways Project (SPP, RRID:SCR_018412)
What is the Signaling Pathways Project (SPP)?
Signaling Pathways Project (SPP) is a web-based multi-omics knowledgebase based upon public, manually curated transcriptomic and cistromic archived datasets involving genetic and small molecule manipulations of cellular receptors, enzymes and transcription factors for mammalian cellular signaling pathways. SPP simplifies powerful data mining and hypothesis generation strategies for the bench researcher, putting a 200 million data point-strong universe of 'omics data points at their fingertips.
Why is SPP important to the scientific community?
Although scientists have generated and archived numerous expression profiling and ChIP-Seq datasets relevant to a broad variety of biological and disease contexts, these datasets can be difficult to access and re-use for hypothesis generation. SPP provides a web-based platform that integrates these datasets published in the literature into consensomes. Consensomes are a list of genes ranked according to a meta-analysis of their differential expression in publicly archived transcriptomic datasets involving perturbations of a specific signaling pathway in a given biosample category. Consensomes are intended as a guide to identifying those genes most consistently impacted by a given pathway in a given tissue context. Scientists can use the SPP platform to survey across millions of biocurated 'omics data points to make high-confidence connections between genomic targets, their upstream regulatory pathways, and disease states.
Interested in learning how to use SPP?
SPP step-by-step tutorials at dkNET Hypothesis Center: https://dknet.org/hypothesis-center/tutorials?facet=SCR_018412
SPP live demo presentation slides: https://www.slideshare.net/dkNET/dknet-webinar-dknet-hypothesis-center-signaling-pathways-project-live-demo
Learn how to query and visualize datasets in SPP and NDEx
Follow these step-by-step instructions for data mining of MERS-CoV, SARS-CoV-1, and SARS-COV-2 transcriptional networks using SPP and NDEx.
https://www.youtube.com/watch?v=x1gqI4XRpkw
0' 13" - Strategy 1: SPP Ominer single gene query
4' 13" - Strategy 2: SPP Ominer consensome query
5' 33" - Strategy 3: SPP Ominer GO term query
6' 45" - Strategy 4: SPP Ominer gene list query
8' 00" - Strategy 5: NDEX consensome network
9' 00" - Strategy 6: NDEX HCT overlap networks
Learn how to compute of a gene list against SARS-Cov-2 consensome
Follow these step-by-step instructions:
https://www.youtube.com/watch?v=jqyvHsORCZ0
Learn more about how researchers used SPP to discover unrecognized connections between coronavirus infection and human signaling pathways?
Check out this article published by SPP team in Scientific Data: https://www.nature.com/articles/s41597-020-00628-6
References
Ochsner, S.A., Pillich, R.T. & McKenna, N.J. Consensus transcriptional regulatory networks of coronavirus-infected human cells. Sci Data 7, 314 (2020). https://doi.org/10.1038/s41597-020-00628-6
Baylor College of Medicine News: https://blogs.bcm.edu/2020/10/13/from-the-labs-web-resources-make-covid-19-research-more-accessible-to-the-scientific-community/





