Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
 MARRVEL(Model organism Aggregated Resources for Rare Variant ExpLoration) Tutorial
MARRVEL(Model organism Aggregated Resources for Rare Variant ExpLoration) Tutorial
Introduction:
MARRVEL is a web tool to search multiple public variant databases simultaneously and provide a unified interface to facilitate the search process. It is used for integration of human and model organism genetic resources to facilitate functional annotation of the human genome. It is used for analysis of human genes and variants by cross-disciplinary integration of records available in public databases to facilitate clinical diagnosis and basic research.
dkNET Webinar: Solving the Undiagnosed Diseases through Machine Learning
In this webinar, Undiagnosed Diseases Network (UDN) project is introduced by Dr. Zhandong Liu. You will learn about MARRVEL, a systematic and comprehensive search engine, and MARRVEL-AI, a knowledge-based and explainable artificial intelligent system to prioritize and identify novel disease-causing coding variants.
Tutorial That Has Been Selected For You:
|
What hypothesis are you developing or what information are you looking for? Evidence for a variant or a gene that is likely to be pathogenic in human diseases |
The hypothesis generation examples that you selected: Pathogenic evidence in human diseases for the mouse gene Acat1 |
Learning Goals:
Learn how to do MARRVEL queries based on the gene or variant of interest
Learn how to navigate the results
Learn how to interpret the results from OMIM and other databases and determine whether this gene is likely to be pathogenic via MARRVEL
Tutorial Lesson:
BACKGROUND: MOUSE GENES THAT ARE INVOLVED IN INCREASED BODY WEIGHT
Acat1 is one of mouse genes that is involved in increased body weight. Here we are interested in finding out whether Acat1 is likely to be pathogenic in human disease using MARRVEL. If you are interested in identifying a gene list in association with mouse phenotypes, you can use the tool provided by MMPC (Mouse Metabolic Phenotyping Centers). A step-by-step tutorial of MMPC is provided in dkNET Hypothesis Center: https://dknet.org/hypothesis-center/tutorials/MMPC?id=7
In the MMPC results page, you will find a list of mouse genes that are associated with the phenotype “increased body weight”. In this example, there are 42 genes returned. Clicking the text 42 Gene Returns will expand the list of genes. You can find that Acat1 is one of the genes listed. You can also check if other genes that are involved in increased body weight from the list are likely to be pathogenic via MARRVEL using the same tutorial below.

STEP 1: Go to MARRVEL
To find whether Acat1 is likely to be pathogenic in human disease using MARRVEL, first let’s open a new window with the MARRVEL
To make it easier to walk through the tutorial, please launch a new window
STEP 2: Since the gene Acat1 that you found from MMPC is a mouse gene, click “Model Organism Gene”. Now you can select the model organism of interest “Mouse (Mus musculus), and input gene symbol. Here we input Acat1 and then click search.
MARRVEL provides information from human genetics databases (OMIM, ExAC, gnomAD, Geno2MP, ClinVar, DGV, and DECIPHER), key eukaryotic genetic model organism and gene ontology databases (SGD, PomBase, WormBase, FlyBase, ZFin, MGI, RGD, GO Central), and others (DIOPT, dbNSFP, Mutalyzer, TransVar, GTEx, Human Protein Atlas). For complete list, please check this paper (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6750039/pdf/nihms-1037046.pdf) or MARRVEL website.

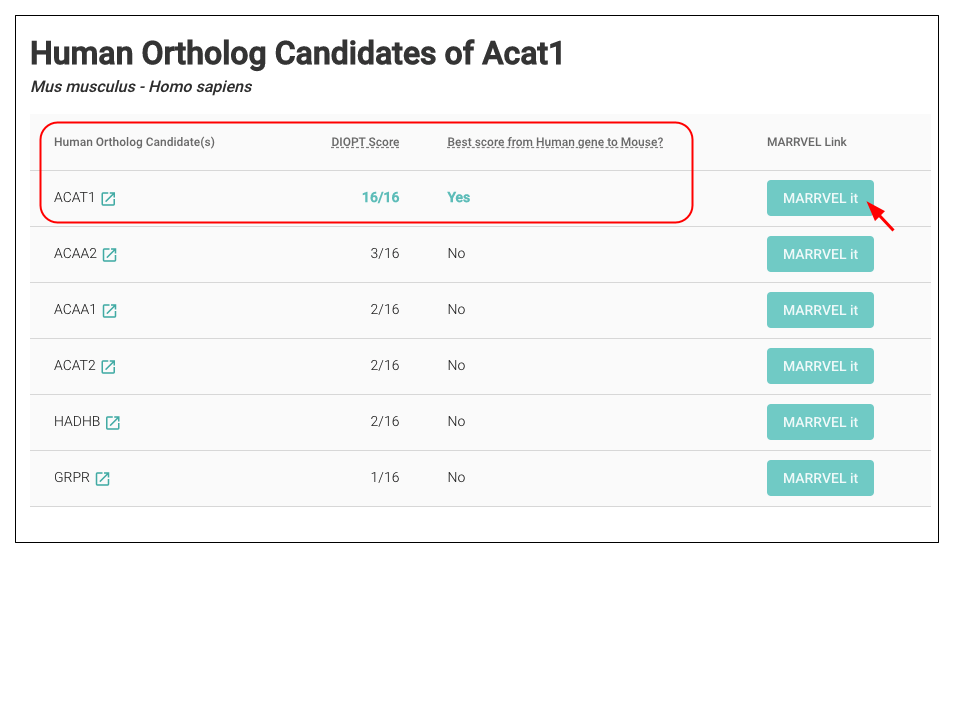
STEP 3: On the next page, you will see a list of predicted human ortholog of choice. Select the one of your interest. Based on the DIOPT score provided from MARRVEL, we will select the first candidate with DIOPT Score 16/16. Now you can click the “MARRVEL it” button.
Which human ortholog should I select?
DIOPT Score is a score of the number of individual ortholog prediction tools that report a given ortholog pair, or that predict the gene as an ortholog of the human gene of interest. The maximum score depends on the number of ortholog prediction tools that include that species in analysis. The “Best score from Human gene to model organism (Mouse)?” helps the selection of the human gene that are likely to be true orthologs of the model organism gene of interest. When “Best score from Human gene to model organism?” is “Yes,” it indicates that the human gene is more likely to be a true human ortholog of the gene of interest. Note that due to evolutionary genome duplications and other phenomenon, the relationships between orthologs are often not 1:1 and are often 1:2 or 1:4, etc.
STEP 4: Now you will see the results page containing a list of data from all the databases that MARRVEL queries. Click on a database from the menu on the right to navigate the results.
How to interpret the results?
Check the data from OMIM (Online Mendelian Inheritance in Man) can help you to determine if your gene of interest is associated with a human disease.
-
What is OMIM?
OMIM (https://www.omim.org) is an online catalog of human genes and genetic disorders, for clinical features, phenotypes and genes. It contains a collection of human genes and genetic phenotypes, focusing on the relationship between phenotype and genotype. Referenced overviews in OMIM contain information on all known mendelian disorders and a variety of related genes. It is updated daily, and entries contain copious links to other genetics resources. -
Learn more about OMIM:
https://www.omim.org/static/omim/pdf/Amberger_at_al_2019_Nucl_Acids_Res.pdf https://www.omim.org/static/omim/pdf/Amberger_at_al_2015_Nucl_Acids_Res.pdf
Under the “OMIM Description of Gene Name” section (in this example, OMIM description of ACAT1), you can find a short summary of what is known about the gene and gene product.
Under the “Disease/Phenotype Associated with Gene Name curated by OMIM ” box (in this example, Disease/Phenotype Associated with ACAT1 curated by OMIM), you can find out if this gene is a known phenotype-associated gene or not. In this case, it shows that ACAT1 is associated with the phenotype Alpha-methyl acetoacetic aciduria and the inheritance is autosomal recessive. You can click the green text “ more on OMIM” to find out more information about this Gene-Phenotype relationship such as cytogenetic location provided by OMIM (OMIM webpage example for the gene ACAT1 https://www.omim.org/entry/607809)
Under the “Reported Alleles with Gene Name curated by OMIM” section (in this example, Reported Alleles with ACAT1 curated by OMIM), you will find a list of pathogenic variants curated by OMIM. Note that OMIM may still miss recently reported disease associations. Thus, we recommend users to conduct PubMed searches as well. In this example, 16 Alleles with mutation of ACAT1 are found with the phenotype 3-@Ketothiolase deficiency. A list of short genetic variations (SNPs) are shown. Clicking the text SNPs (i.e.,in this example, rs120074140) will bring you to the NLM NCBI dbSNP Reference SNP Report, where you can find more detailed information about a specific SNP (see an example of SNP report of rs120072140 https://www.ncbi.nlm.nih.gov/snp/rs120074140?horizontal_tab=true)
Please note that some data will not be displayed because a variant is not entered when starting with a model organism gene. Some data are only displayed when a variant is entered. For example, the allele frequency from gnomAD, ExAC, and Geno2MP won’t be displayed without variant information. The pathogenicity prediction algorithms will also not be available. For Geno2MP, a summary of the types of variants present in this database will be displayed instead. For ClinVar, there will be no variants highlighted.

Here we only give you an example of how to interpret the results from OMIM. MARVELL provides query results from the following databases (OMIM, ExAC, gnomAD, Geno2MP, ClinVar, DGV, and DECIPHER). You can click the database name from the right side panel and see the results from other databases.
-
Where is the MARRVEL data aggregated from?
Check out more information at MARRVEL website: http://marrvel.org/faq -
ClinVar: It is an archive of aggregated information about sequence variation and its relationship to human health. It provides reports of relationships among human variations and phenotypes along with supporting evidence. Submissions from clinical testing labs, research labs, locus-specific databases, expert panels and professional societies are welcome. It collects reports of variants found in patient samples, assertions made regarding their clinical significance, information about submitter, and other supporting data. Alleles described in submissions are mapped to reference sequences, and reported according to HGVS standard. For more information on ClinVar please see: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4832236/
-
Geno2MP: It is a web-based query tool that searches a database of rare variants from exome sequencing data linked to phenotypic information from a wide variety of Mendelian gene discovery projects. It is a collection of phenotypic profiles for affected individuals and, for unaffected individuals, the phenotypic profile of their affected relative(s). It is a collaborative, shared resource for the human genetics community. For more information on Geno2MP please see: https://geno2mp.gs.washington.edu/Geno2MP/#/instructions
-
DECIPHER: It is an Interactive database which incorporates a suite of tools designed to aid the interpretation of submicroscopic chromosomal imbalance. It is used to enhance clinical diagnosis by retrieving information from bioinformatics resources relevant to the imbalance found in the patient. Users can find common copy-number variants and their frequencies from MARRVEL. For more information on DECIPHER please see: https://www.deciphergenomics.org/about/help
-
GnomAD: A large population genomics databases based on Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS) of people who are selected to exclude severe pediatric diseases. GnomAD provides a control population frequency of variants (123,136 exomes and 15,496 genomes). Cohorts are selected to exclude severe pediatric diseases. For more information on GnomAD please see: https://arxiv.org/abs/2107.11458
-
DGV: It is a collection of curated structural variation in the human genome. Catalogue of human genomic structural variation identified in healthy control samples for studies aiming to correlate genomic variation with phenotypic data. It is continuously updated with new data from peer reviewed research studies. For more information on DGV please see: http://dgv.tcag.ca/dgv/app/resources?ref=GRCh37/hg19
Reference
Wang J, Mao D, Fazal F, Kim SY, Yamamoto S, Bellen H, Liu Z. Using MARRVEL v1.2 for Bioinformatics Analysis of Human Genes and Variant Pathogenicity. Curr Protoc Bioinformatics. 2019 Sep;67(1):e85. doi: 10.1002/cpbi.85. PMID: 31524990; PMCID: PMC6750039.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6750039/pdf/nihms-1037046.pdf




