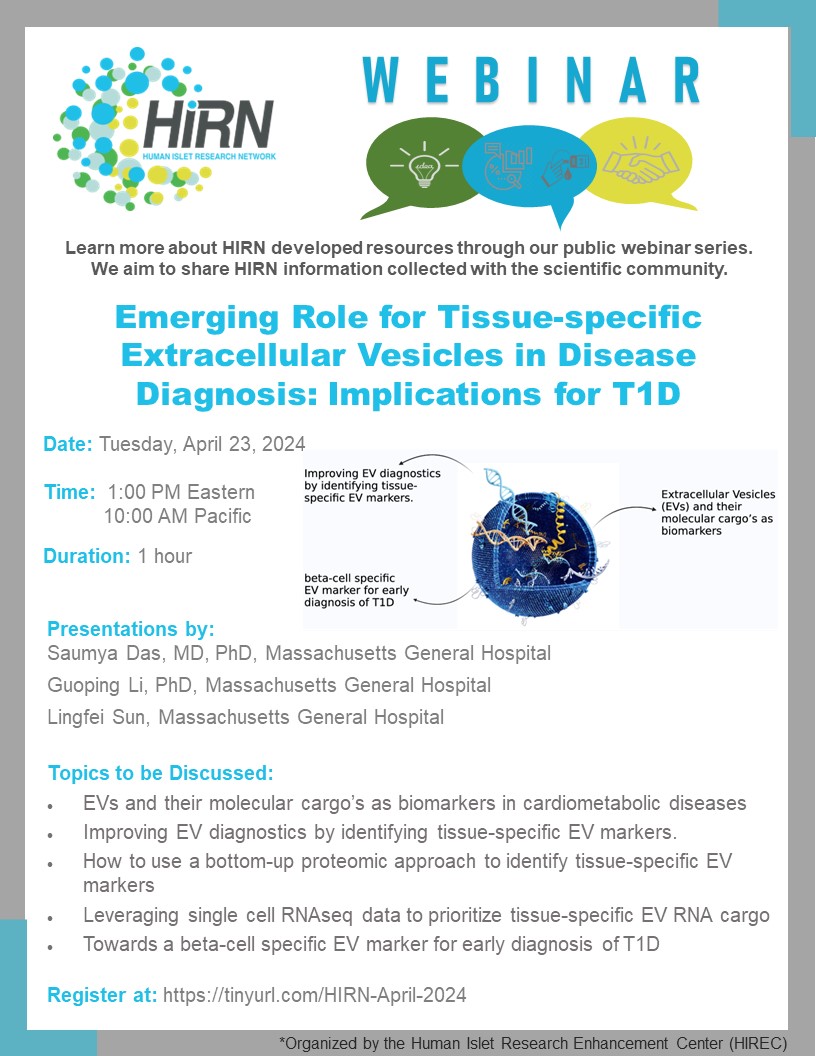
Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
December news from our collaborating resource center - NURSA
Here is the content published by NURSA on December 1, 2016:

|
In this month's NURSA Signals: a call for papers for a new special issue of Molecular and Cellular Endocrinology, 'Omics of Nuclear Receptor Signaling, edited by Neil McKenna; a special focus on adipose & obesity datasets; integration with GeneCards; and some exciting plans for the New Year.
| Molecular and Cellular Endocrinology Special Issue: 'Omics of Nuclear Receptor Signaling: Call for Papers |
Does your lab have an unpublished expression profiling, ChIP-Seq, proteomic or metabolomic dataset related to nuclear receptor or coregulator signaling that you would like to publish but aren't sure where? The NURSA website editor Neil McKenna is editing a special issue of Molecular and Cellular Endocrinology, entitled "'Omics of Nuclear Receptor Signaling" that will feature articles based upon 'omics/discovery-scale datasets. Unlike traditional research manuscripts, there will be less emphasis in these articles on deep mechanistic validation and contextualization, and more on the dataset itself as an environment for shedding light on novel or uncharacterized biology. Uniquely, these articles will be linked into NURSA's steadily expanding library of pan-omics datasets to extend and strengthen their biological impact.
See the MCE website announcement for more details. Please note that the submission deadline has been extended to February 28, 2017.
| NURSA Datasets: Focus on Adipose Tissue |
Insights into the roles played by nuclear receptors and other signaling pathways in adipose biology are supported by numerous articles based in part upon transcriptomic datasets. In advance of a NURSA presence at the forthcoming Keystone Obesity and Adipose Tissue Biology meeting in Keystone, Colorado in January 2017, our biocuration team is currently focused on datasets containing transcriptomic signatures of obesity and adipose signaling by nuclear receptors and other pathways.
Recent examples include:
Analysis of the dexamethasone (Dex)-regulated transcriptome in human subcutaneous preadipocytes
Filter on "Metabolic.....Adipose" in the Biosource dropdown in the NURSA Dataset Directory for the full listing and look out for future dataset releases.
| GeneCards integration |
We're pleased to announce that the 500 lb gorilla of curated gene-level information, GeneCards, now includes links to Transcriptomine Regulation Reports in the "Expression" subsection of its gene pages. Check out the link to the Transcriptomine GREB1 Regulation Report in the GeneCard GREB1 page. Our thanks to Genecards' Doron Lancet and Marilyn Szafran for making this happen.
| Plans for 2017 |
Web development
The NURSA web development team has been hard at work over the summer preparing for a major new release of the site in the early New Year, which will include improvements to the Transcriptomine search results and dataset pages. Specifically, we'll be introducing cut-offs and a fold-change slider that will optimize the visual organization of the search results and prevent blow-out fold changes (PPARG pathway induction of FABP4 in adipose tissue we're looking at you) from skewing the charts.
A new categorization system in gene Regulation Reports will highlight gene fold changes in animal & cell model systems (think bile duct ligation or adipogenesis in 3T3-L1 cells), clinical datasets and datasets involving perturbations in non-NR pathways like CMET. So if a gene is highly regulated by the glucocorticoid pathway as well as during inflammatory responses and in adipose tissue from obese individuals, you'll know about it at a glance using Transcriptomine. We're also introducing scatterplots to the dataset pages to allow for a quick visual appreciation of the top regulated genes in each experiment.
Biocuration
On the biocuration side, we'll be filling a couple of new biocuration positions in the New Year that will catalyze our efforts to add cistromic (ChIP-Seq) datasets to the NURSA dataset library later in 2017. Finally, we're bound to silence for now, but plans are afoot for an exciting collaboration with a major publisher that will place 'omics-scale datasets in the cell signaling field on a whole new level of accessibility and re-usability.
Stay tuned!
Sincerely
The NURSA team
|